Help
1 Overview
Pyrus Genome Database collects genome and transcriptome data of pear accessions, and provides functional modules for easy searching, analysis and visualization. Users can quickly obtain desired functional information for biological discovery.
2 Search
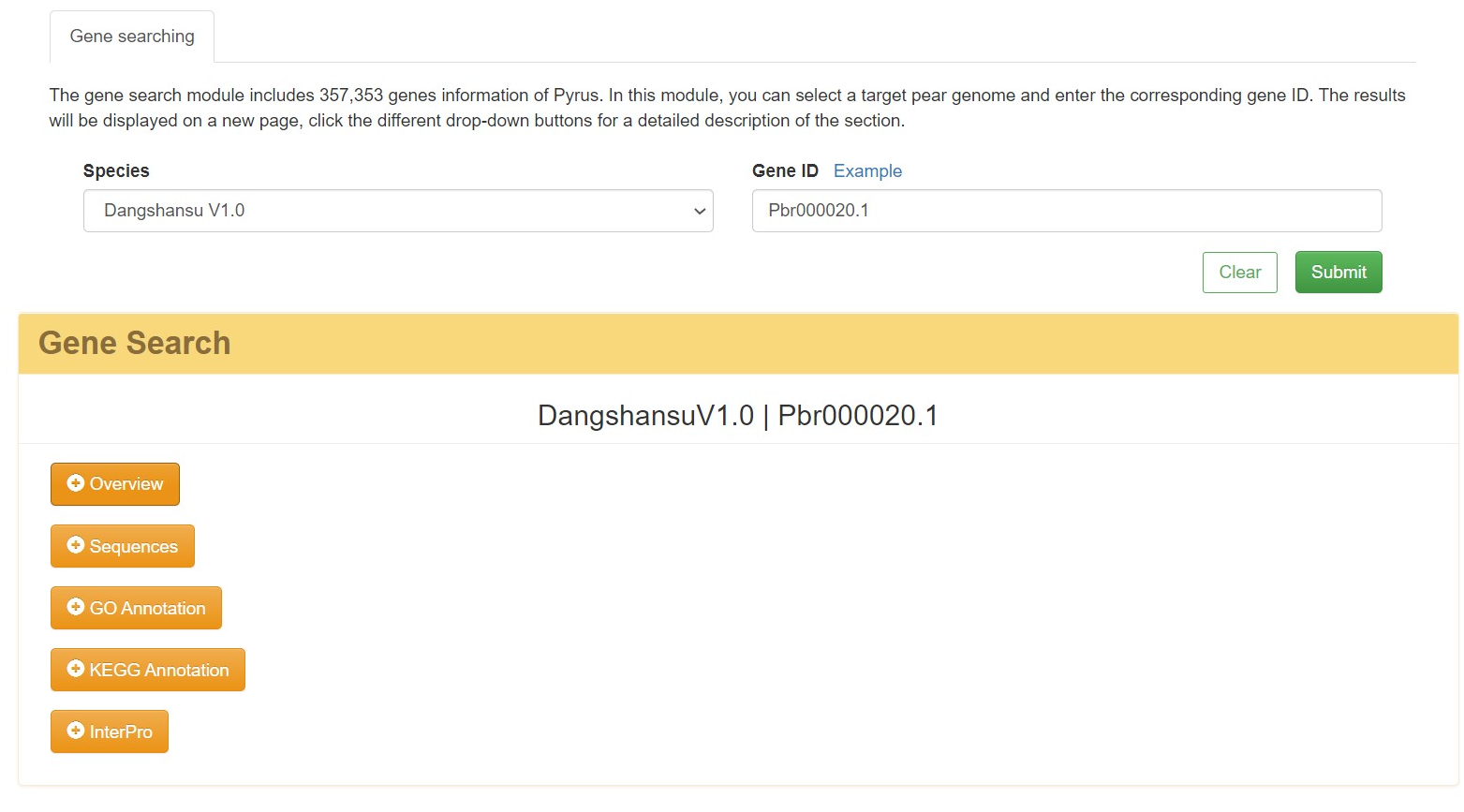
2.1 Quick search-Gene information
This module includes 357,353 genes of Pyrus. In this module, simply select a target pear genome, type select gene, and enter the corresponding gene ID. The results will be displayed on a new page, and users can click the different drop-down buttons for a detailed description of the section. It includes the basic information of the genes, three types of sequence information (gene, mRNA, protein sequence) , functional annotation information (GO, KEGG, interpro) and homologous analysis results.
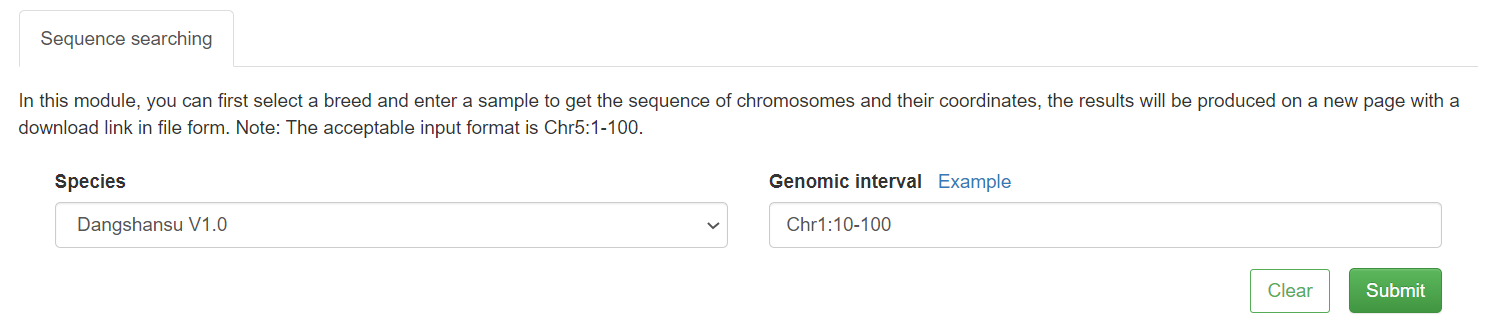
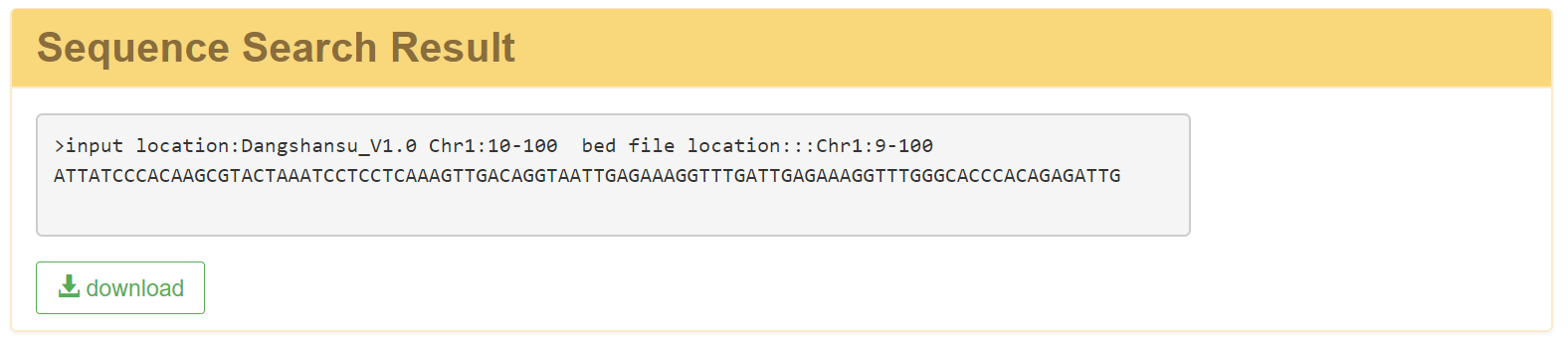
2.2 Quick search-Genome sequence
In this module, just simply select a target pear genome, type select genome, and enter the position coordinates.The related sequences will be stored in a file, and generated on a new page with a download link.
2.3 Sequence fetch
Select a target pear genome and the desired sequence type, and enter a list of ids (note: one ID per line). The results will be presented in a new page with a download link.


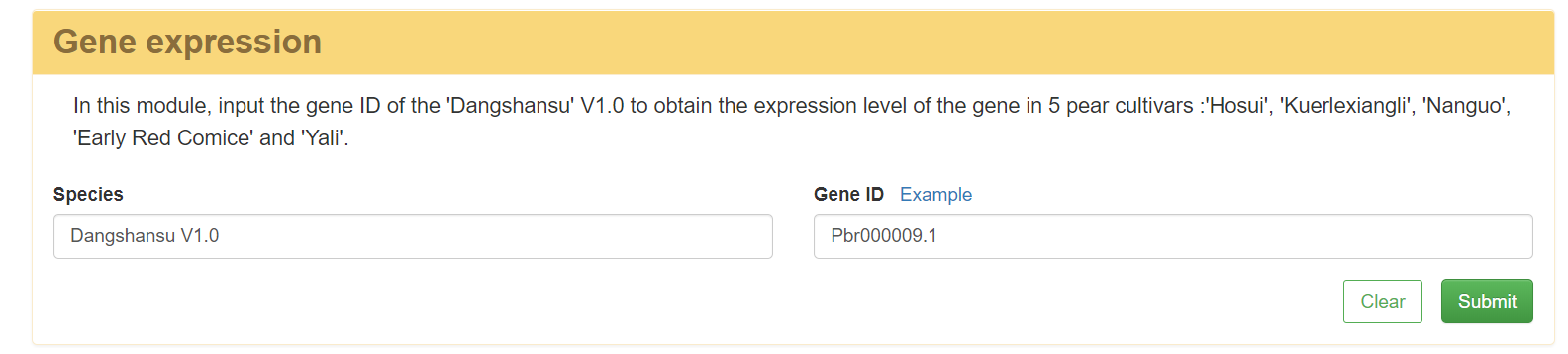
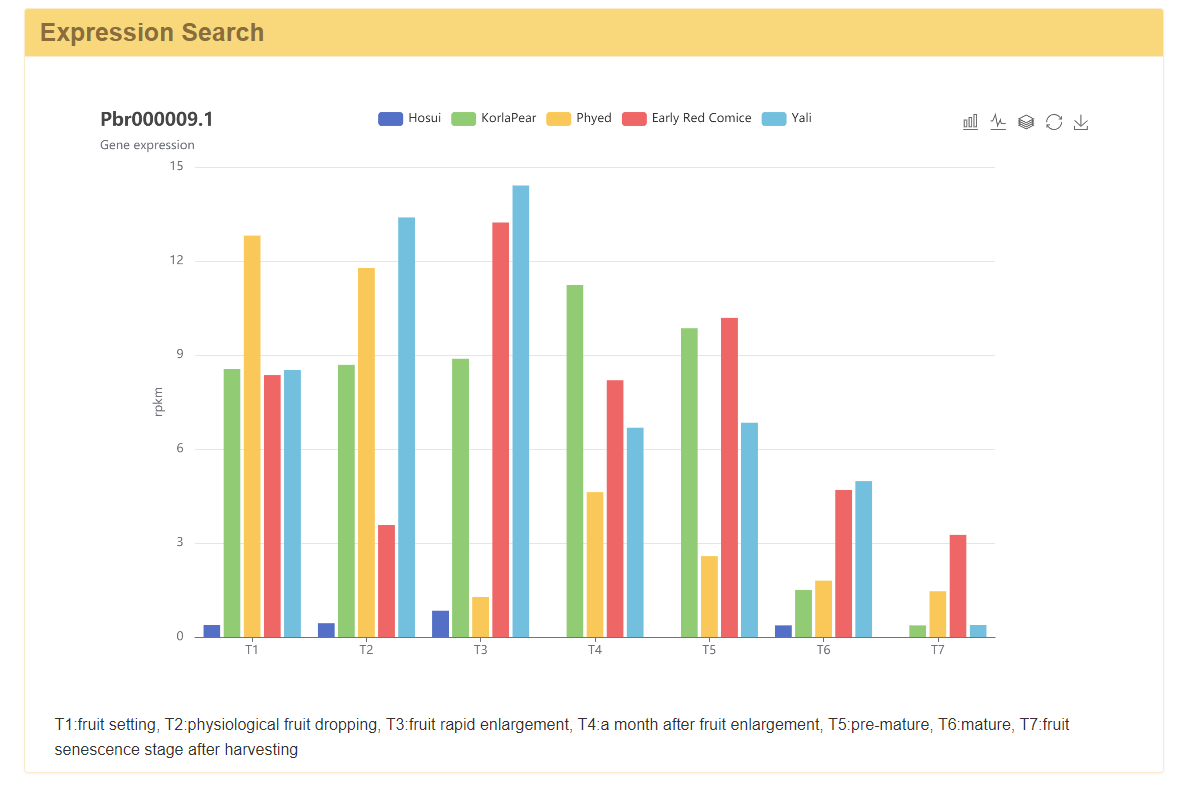
3 Gene expression
In this module, gene expression of five pear cultivars ('Hosui', 'Kuerlexiangli', 'Nanguo', 'Early Red Comice', and 'Yali') in 7 periods were obtained. The gene expression level of pear cultivars will be diagramed by inputting the 'Dangshansuli' V1.0 gene ID.
4 Blast
4.1 Basic blast
| Query Type | Database Type | BLAST Program |
|---|---|---|
| Nucleotide | Nucleotide | blastn: Search a nucleotide database using a nucleotide query. |
| tblastx: Search translated nucleotide database using a translated nucleotide query. | ||
| Protein | blastx: Search protein database using a translated nucleotide query. | |
| Protein | Nucleotide | tblastn: Search translated nucleotide database using a protein query. |
| Protein | blastp: Search protein database using a protein query. |
4.2 Advanced blast
5 Synteny
In this module, we provide synteny block Search for Oriental pear('Dangshansuli' V1.0) and Occidental pear (Bartlett V2.0).
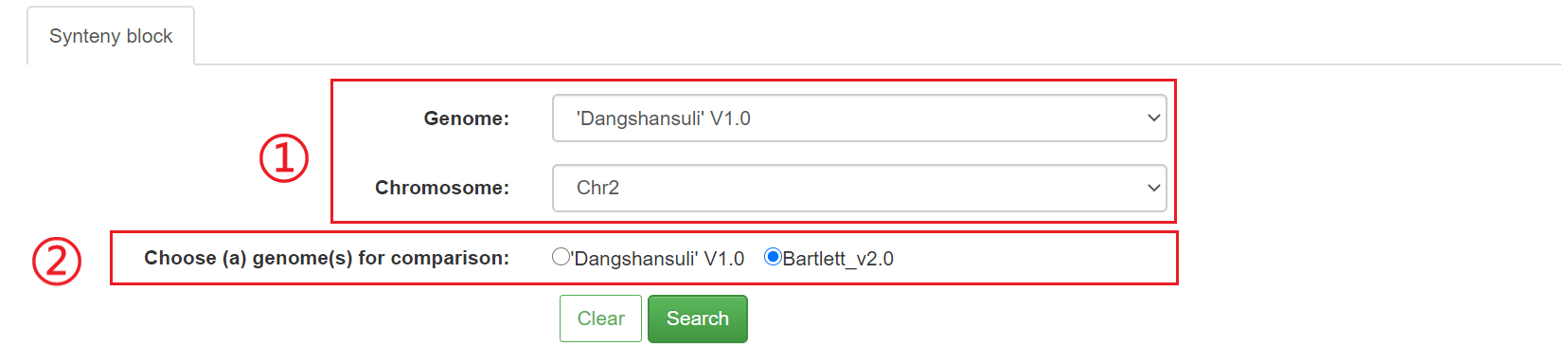
5.1 Synteny block
In this module, MCScanX and BlastP were used to analyze conserved syntenic blocks between the Oriental pear ('Dangshansuli' V1.0) and the Occidental pear (Bartlett V2.0).
Steps for usage:
1) Select query genome 'A' and a specific chromosome 'a'
2) Select the genome 'B' that needs to be compared
3) Click the Submit button
4) The results will be divided into three parts:
① The image shows the number of conserved syntenic blocks between chromosome 'a' and genome 'B'.
② The table shows the detailed location and matching of each syntenic block.
③ Clicking on each synteny block links to detailed genetic information in the block.
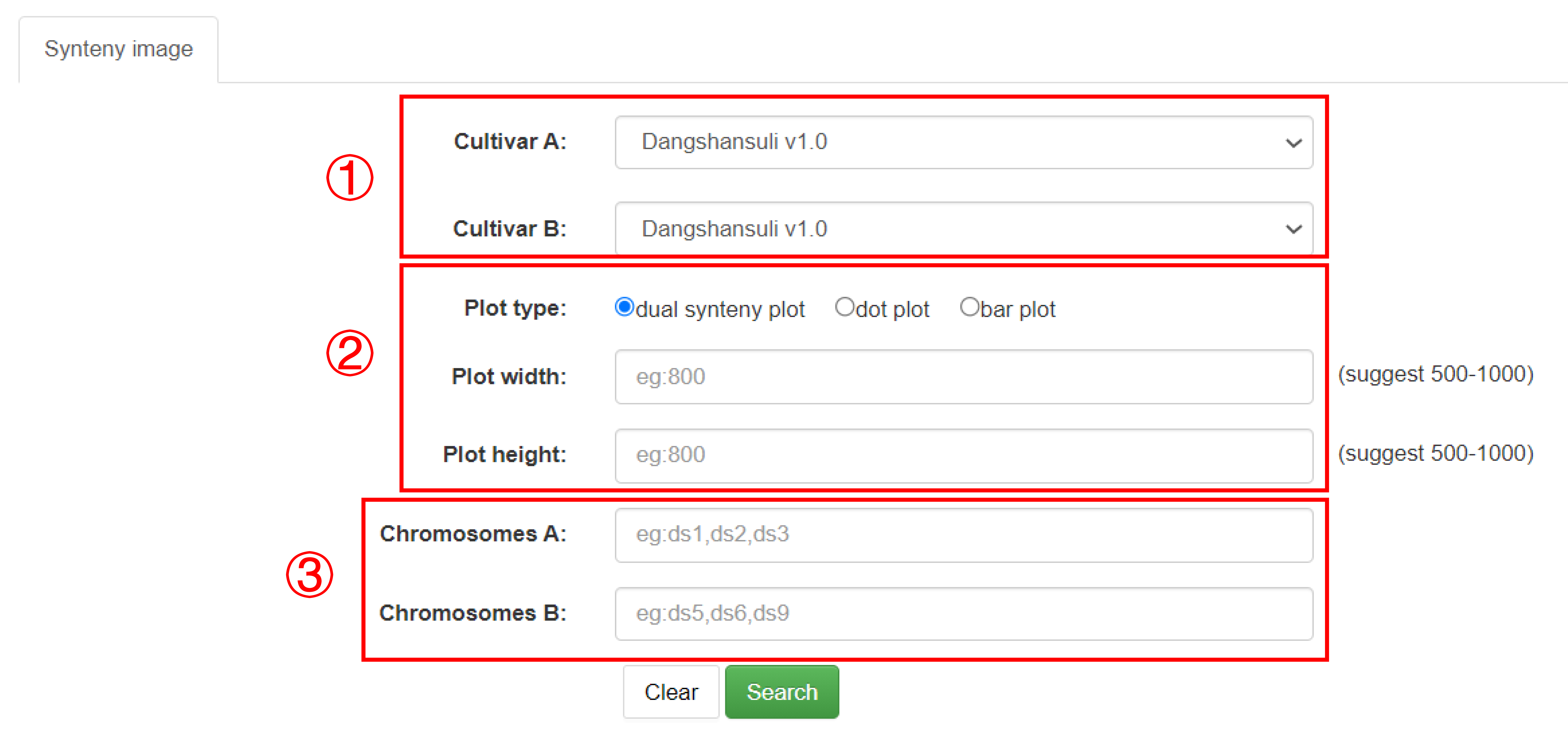
5.2 Synteny image
In this module, MCScanX and BlastP were used to provide the ability to plot three types of images (Synteny plot, dot plot and bar plot) online between any two chromosomes of two genomes.
Steps for usage:
1) Select the genomes 'A' or 'B'.
2) Fill in the image parameters: Plot type, Plot width, and Plot height.
3) Fill in the specific chromosomes to be drawn in the two genomes, respectively. ('Dangshansuli' is represented by 'ds' and Bartlett_V2.0 is represented by 'bt').
4) Click the Submit button.
5) The results will be presented as an image with download link.
Note:
When drawing a synteny image, specific chromosome ids are required for different genomes.
(For example: Dangshansuliv1.0-ds1, Cuiguanv1.0-cgChr1, Shanxiduliv1.0-sdChr1, Zhongai NO.1-zaChr1, Bartlett v1.0-Btscaffold00001, Bartlett v2.0-bt00000039, Nijisseiki-niChr1, d'Anjou-ajDAnjou_Chr0, Dangshansuliv1.1-DsChr1)
6 Phylogenetic Tree Build
This module uses mafft and Iqtree to provide the Phylogenetic Tree online.
Two result files will be provided:
1) Perform multiple sequence alignments on submitted sequences:result.mafft
2) Phylogenetic Tree file in NWK format
Tips:
At least 4 sequences are required,eg:
>name
ATGCATGC
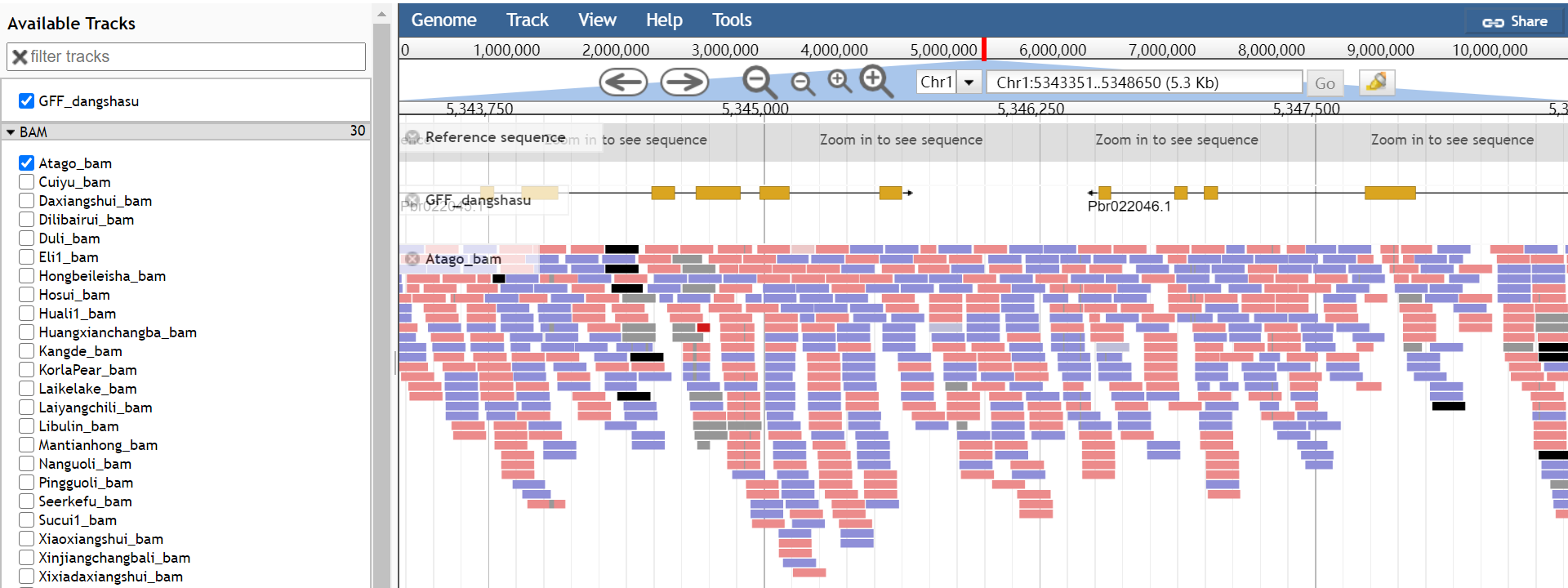
7 JBrowse
PGDB provides JBrowse function for eight pear genomes. In addition, the SNP data of 30 pear accessions were also included in Jbrowse- 'Dangshansuli' V1.0.